vegas Module¶
Introduction¶
The key Python objects and functions supported by the vegas module are:
vegas.Integrator— an object describing a multidimensional integration operator. Such objects contain information about the integration volume, and also about optimal remappings of the integration variables based upon the last integral evaluated using the object.
vegas.AdaptiveMap— an object describing the remappings used byvegas.
vegas.RAvg— an object describing the result of avegasintegration.vegasreturns the weighted average of the integral estimates from eachvegasiteration as an object of classvegas.RAvg. These are Gaussian random variables — that is, they have a mean and a standard deviation — but also contain information about the iterationsvegasused in generating the result.
vegas.RAvgArray— an array version ofvegas.RAvgused when the integrand is array-valued.
vegas.RAvgDict— a dictionary version ofvegas.RAvgused when the integrand is dictionary-valued.
vegas.lbatchintegrand()— a decorator that labels a function as an lbatch integrand.
vegas.rbatchintegrand()— a decorator that labels a function as an rbatch integrand.
vegas.PDFIntegrator— a specialized integrator for evaluating expectation values with arbitrary probability density functions.
vegas.PDFEV— an expectation value fromvegas.PDFIntegrator.
vegas.PDFEVArray— an array of expectation values fromvegas.PDFIntegrator.
vegas.PDFEVDict— a dictionary of expectation values fromvegas.PDFIntegrator.
These are described in detail below.
Integrator Objects¶
The central component of the vegas package is the integrator class:
- class vegas.Integrator¶
Adaptive multidimensional Monte Carlo integration.
vegas.Integratorobjects make Monte Carlo estimates of multidimensional functionsf(x)wherex[d]is a point in the integration volume:integ = vegas.Integrator(integration_region) result = integ(f, nitn=10, neval=10000)
The integator makes
nitnestimates of the integral, each using at mostnevalsamples of the integrand, as it adapts to the specific features of the integrand. Successive estimates (iterations) typically improve in accuracy until the integrator has fully adapted. The integrator returns the weighted average of allnitnestimates, together with an estimate of the statistical (Monte Carlo) uncertainty in that estimate of the integral. The result is an object of typeRAvg(which is derived fromgvar.GVar).Integrands
f(x)return numbers, arrays of numbers (any shape), or dictionaries whose values are numbers or arrays (any shape). Each number returned by an integrand corresponds to a different integrand. When arrays are returned,vegasadapts to the first number in the flattened array. When dictionaries are returned,vegasadapts to the first number in the value corresponding to the first key.vegascan generate integration points in batches for integrands built from classes derived fromvegas.LBatchIntegrand, or integrand functions decorated byvegas.lbatchintegrand(). Batch integrands are typically much faster, especially if they are coded in Cython or C/C++ or Fortran.vegas.Integrators have a large number of parameters but the only ones that most people will care about are: the numbernitnof iterations of thevegasalgorithm; the maximum numbernevalof integrand evaluations per iteration; and the damping parameteralpha, which is used to slow down the adaptive algorithms when they would otherwise be unstable (e.g., with very peaky integrands). Setting parameteranalyzer=vegas.reporter()is sometimes useful, as well, since it causesvegasto print (onsys.stdout) intermediate results from each iteration, as they are produced. This helps when each iteration takes a long time to complete (e.g., longer than an hour) because it allows you to monitor progress as it is being made (or not).- Parameters:
map (array, dictionary,
vegas.AdaptiveMaporvegas.Integrator) –The integration region is specified by an array
map[d, i]wheredis the direction andi=0,1specify the lower and upper limits of integration in directiond. Integration pointsxare packaged as arraysx[d]when passed to the integrandf(x).More generally, the integrator packages integration points in multidimensional arrays
x[d1, d2..dn]when the integration limits are specified bymap[d1, d2...dn, i]` with ``i=0,1. These arrays can have any shape.Alternatively, the integration region can be specified by a dictionary whose values
map[key]are either 2-tuples or arrays of 2-tuples corresponding to the lower and upper integration limits for the corresponding variables. Then integration pointsxdare packaged as dictionaries having the same structure asmapbut with the integration limits replaced by the values of the variables: for example,map = dict(r=(0, 1), phi=[(0, np.pi), (0, 2 * np.pi)])
indicates a three-dimensional integral over variables
r(from0to1),phi[0](from0tonp.pi), andphi[1](from0to2*np.pi). In this case integrandsf(xd)have dictionary argumentsxdwherexd['r'],xd['phi'][0], andxd['phi'][1]correspond to the integration variables.Finally
mapcould be the integration map from anothervegas.Integrator, or thatvegas.Integratoritself. In this case the grid is copied from the existing integrator.uses_jac (bool) – Setting
uses_jac=Truecausesvegasto call the integrand with two arguments:fcn(x, jac=jac). The second argument is the Jacobianjac[d] = dx[d]/dy[d]of thevegasmap. The integral overx[d]of1/jac[d]equals 1 (exactly). The default setting isuses_jac=False.nitn (positive int) – The maximum number of iterations used to adapt to the integrand and estimate its value. The default value is 10; typical values range from 10 to 20.
neval (positive int) – Approximate number of integrand evaluations in each iteration of the
vegasalgorithm. Increasingnevalincreases the precision: statistical errors should fall at least as fast assqrt(1./neval)and often fall much faster. The default value is 1000; real problems often require 10–10,000 times more evaluations than this.nstrat (int array) –
nstrat[d]specifies the number of stratifications to use in directiond. By default this parameter is set automatically, based on parameterneval, withnstrat[d]approximately the same for everyd. Specifyingnstratexplicitly makes it possible to concentrate stratifications in directions where they are most needed. Ifnstratis set butnevalis not,nevalis set equal to2*prod(nstrat)/(1-neval_frac).alpha (float) – Damping parameter controlling the remapping of the integration variables as
vegasadapts to the integrand. Smaller values slow adaptation, which may be desirable for difficult integrands. Small or zeroalphas are also sometimes useful after the grid has adapted, to minimize fluctuations away from the optimal grid. The default value is 0.5.beta (float) – Damping parameter controlling the redistribution of integrand evaluations across hypercubes in the stratified sampling of the integral (over transformed variables). Smaller values limit the amount of redistribution. The theoretically optimal value is 1; setting
beta=0prevents any redistribution of evaluations. The default value is 0.75.neval_frac (float) – Approximate fraction of function evaluations used for adaptive stratified sampling.
vegasdistributes(1-neval_frac)*nevalintegrand evaluations uniformly over all hypercubes, with at least 2 evaluations per hypercube. The remainingneval_frac*nevalevaluations are concentrated in hypercubes where the errors are largest. Increasingneval_fracmakes more integrand evaluations available for adaptive stratified sampling, but reduces the number of hypercubes, which limits the algorithm’s ability to adapt. Ignored whenbeta=0. Default isneval_frac=0.75.adapt (bool) – Setting
adapt=Falseprevents further adaptation byvegas. Typically this would be done after training thevegas.Integratoron an integrand, in order to stabilize further estimates of the integral.vegasuses unweighted averages to combine results from different iterations whenadapt=False. The default setting isadapt=True.nproc (int or None) –
Number of processes/processors used to evalute the integrand. If
nproc>1Python’smultiprocessingmodule is used to spread integration points across multiple processors, thereby potentially reducing the time required to evaluate the integral. There is a significant overhead involved in using multiple processors so this option is useful only when the integrand is expensive to evaluate. When using themultiprocessingmodule in its default mode for MacOS and Windows, it is important that the main module can be safely imported (i.e., without launching new processes). This can be accomplished with some version of theif __name__ == '__main__`:construct in the main module: e.g.,if __name__ == '__main__': main()
This is not an issue on other Unix platforms. See the
multiprocessingdocumentation for more information. Note that settingnprocgreater than 1 disables MPI support. Setnproc=Noneto use all the processors on the machine (equivalent tonproc=os.cpu_count()). Default value isnproc=1. (Requires Python 3.3 or later.)Note that
nprochas nothing to do with MPI support. The number of MPI processors is specified outside Python (via, for example,mpirun -np 8 python script.pyon the command line).analyzer –
An object with methods
analyzer.begin(itn, integrator)analyzer.end(itn_result, result)where:
begin(itn, integrator)is called at the start of eachvegasiteration withitnequal to the iteration number andintegratorequal to the integrator itself; andend(itn_result, result)is called at the end of each iteration withitn_resultequal to the result for that iteration andresultequal to the cummulative result of all iterations so far. Settinganalyzer=vegas.reporter(), for example, causes vegas to print out a running report of its results as they are produced. The default isanalyzer=None.min_neval_batch (positive int) – The minimum number of integration points to be passed together to the integrand when using
vegasin batch mode. The default value is 50,000. Larger values may be lead to faster evaluations, but at the cost of more memory for internal work arrays. The last batch is usually smaller than this limit, as it is limited byneval.max_neval_hcube (positive int) – Maximum number of integrand evaluations per hypercube in the stratification. The default value is 50,000. Larger values might allow for more adaptation (when
beta>0), but also allow for more over-shoot when adapting to sharp peaks. Larger values also can result in large internal work arrasy.minimize_mem (bool) – When
True,vegasminimizes internal workspace by moving some of its data to a disk file. This increases execution time (slightly) and results in temporary files, but might be desirable when the number of evaluations is very large (e.g.,neval=1e9).minimize_mem=Truerequires theh5pyPython module.max_mem (positive float) – Maximum number of floats allowed in internal work arrays (approx.). A
MemoryErroris raised if the work arrays are too large, in which case one might want to reducemin_neval_batchormax_neval_hcube, or setminimize_mem=True(or increasemax_memif there is enough RAM). Default value is 1e9.maxinc_axis (positive int) – The maximum number of increments per axis allowed for the x-space grid. The default value is 1000; there is probably little need to use other values.
rtol (float) – Relative error in the integral estimate at which point the integrator can stop. The default value is 0.0 which turns off this stopping condition. This stopping condition can be quite unreliable in early iterations, before
vegashas converged. Use with caution, if at all.atol (float) – Absolute error in the integral estimate at which point the integrator can stop. The default value is 0.0 which turns off this stopping condition. This stopping condition can be quite unreliable in early iterations, before
vegashas converged. Use with caution, if at all.ran_array_generator – Replacement function for the default random number generator.
ran_array_generator(size)should create random numbers uniformly distributed between 0 and 1 in an array whose dimensions are specified by the integer-valued tuplesize. Settingran_array_generatortoNonerestores the default generator (fromgvar).sync_ran (bool) – If
True(default), the default random number generator is synchronized across all processors when using MPI. IfFalse,vegasdoes no synchronization (but the random numbers should synchronized some other way). Ignored if not using MPI.adapt_to_errors (bool) –
adapt_to_errors=Falsecausesvegasto remap the integration variables to emphasize regions where|f(x)|is largest. This is the default mode.adapt_to_errors=Truecausesvegasto remap variables to emphasize regions where the Monte Carlo error is largest. This might be superior when the number of the number of stratifications (self.nstrat) in the y grid is large (> 100). It is typically useful only in one or two dimensions.uniform_nstrat (bool) – If
True, requires that thenstrat[d]be equal for alld. IfFalse(default), the algorithm maximizes the number of stratifications while requiring|nstrat[d1] - nstrat[d2]| <= 1. This parameter is ignored ifnstratis specified explicitly.mpi – Setting
mpi=False(default) disablesmpisupport invegaseven ifmpiis available; settingmpi=Trueallows use ofmpiprovided modulempi4pyis installed.
- Methods:
- __call__(fcn, save=None, saveall=None, **kargs)¶
Integrate integrand
fcn.A typical integrand has the form, for example:
def f(x): return x[0] ** 2 + x[1] ** 4
The argument
x[d]is an integration point, where indexd=0...represents direction within the integration volume.Integrands can be array-valued, representing multiple integrands: e.g.,
def f(x): return [x[0] ** 2, x[0] / x[1]]
The return arrays can have any shape. Dictionary-valued integrands are also supported: e.g.,
def f(x): return dict(a=x[0] ** 2, b=[x[0] / x[1], x[1] / x[0]])
Integrand functions that return arrays or dictionaries are useful for multiple integrands that are closely related, and can lead to substantial reductions in the errors for ratios or differences of the results.
Integrand’s take dictionaries as arguments when
Integratorkeywordmapis set equal to a dictionary. For example, withmap = dict(r=(0,1), theta=(0, np.pi), phi=(0, 2*np.pi))
the volume of a unit sphere is obtained by integrating
def f(xd): r = xd['r'] theta = xd['theta'] return r ** 2 * np.sin(theta)
It is usually much faster to use
vegasin batch mode, where integration points are presented to the integrand in batches. A simple batch integrand might be, for example:@vegas.lbatchintegrand def f(x): return x[:, 0] ** 2 + x[:, 1] ** 4
where decorator
@vegas.lbatchintegrandtellsvegasthat the integrand processes integration points in batches. The arrayx[i, d]represents a collection of different integration points labeled byi=0.... (The number of points is controlledvegas.Integratorparametermin_neval_batch.)Batch mode is particularly useful (and fast) when the integrand is coded in Cython. Then loops over the integration points can be coded explicitly, avoiding the need to use
numpy’s whole-array operators if they are not well suited to the integrand.The batch index is always first (leftmost) for lbatch integrands, as above. It is also possible to create batch integrands where the batch index is the last (rightmost) index: for example,
@vegas.rbatchintegrand def f(x): return x[0, :] ** 2 + x[1, :] ** 4
Batch integrands can also be constructed from classes derived from
vegas.LBatchIntegrandorvegas.RBatchIntegrand.Any
vegasparameter can also be reset: e.g.,self(fcn, nitn=20, neval=1e6).- Parameters:
fcn (callable) – Integrand function.
save (str or file or None) –
Writes
resultsinto pickle file specified bysaveat the end of each iteration. For example, settingsave='results.pkl'means that the results returned by the last vegas iteration can be reconstructed later using:import pickle with open('results.pkl', 'rb') as ifile: results = pickle.load(ifile)
Ignored if
save=None(default).saveall (str or file or None) –
Writes
(results, integrator)into pickle file specified bysaveallat the end of each iteration. For example, settingsaveall='allresults.pkl'means that the results returned by the last vegas iteration, together with a clone of the (adapted) integrator, can be reconstructed later using:import pickle with open('allresults.pkl', 'rb') as ifile: results, integrator = pickle.load(ifile)
Ignored if
saveall=None(default).
- Returns:
Monte Carlo estimate of the integral of
fcn(x)as an object of typevegas.RAvg,vegas.RAvgArray, orvegas.RAvgDict.
- set(ka={}, **kargs)¶
Reset default parameters in integrator.
Usage is analogous to the constructor for
vegas.Integrator: for example,old_defaults = integ.set(neval=1e6, nitn=20)
resets the default values for
nevalandnitninvegas.Integratorinteg. A dictionary, hereold_defaults, is returned. It can be used to restore the old defaults using, for example:integ.set(old_defaults)
- settings(ngrid=0)¶
Assemble summary of integrator settings into string.
- Parameters:
ngrid (int) – Number of grid nodes in each direction to include in summary. The default is 0.
- Returns:
String containing the settings.
- random(yield_hcube=False, yield_y=False)¶
Iterator over integration points and weights.
This method creates an iterator that returns integration points from
vegas, and their corresponding weights in an integral. Each pointx[d]is accompanied by the weight assigned to that point byvegaswhen estimating an integral. Optionally it will also return the index of the hypercube containing the integration point and/or the y-space coordinates:integ.random() yields x, wgt integ.random(yield_hcube=True) yields x, wgt, hcube integ.random(yield_y=True) yields x, y, wgt integ.random(yield_hcube=True, yield_y=True) yields x, y, wgt, hcube
The number of integration points returned by the iterator corresponds to a single iteration.
- random_batch(yield_hcube=False, yield_y=False)¶
Iterator over integration points and weights.
This method creates an iterator that returns integration points from
vegas, and their corresponding weights in an integral. The points are provided in arraysx[i, d]wherei=0...labels the integration points in a batch andd=0...labels direction. The corresponding weights assigned byvegasto each point are provided in an arraywgt[i].Optionally the integrator will also return the indices of the hypercubes containing the integration points and/or the y-space coordinates of those points:
integ.random_batch() yields x, wgt integ.random_batch(yield_hcube=True) yields x, wgt, hcube integ.random_batch(yield_y=True) yields x, y, wgt integ.random_batch(yield_hcube=True, yield_y=True) yields x, y, wgt, hcube
The number of integration points returned by the iterator corresponds to a single iteration. The number in a batch is controlled by parameter
nhcube_batch.
AdaptiveMap Objects¶
vegas’s remapping of the integration variables is handled
by a vegas.AdaptiveMap object, which maps the original
integration variables x into new variables y in a unit hypercube.
Each direction has its own map specified by a grid in x space:
where and
are the limits of integration.
The grid specifies the transformation function at the points
for
:
Linear interpolation is used between those points. The Jacobian for this transformation is:
vegas adjusts the increments sizes to optimize its Monte Carlo
estimates of the integral. This involves training the grid. To
illustrate how this is done with vegas.AdaptiveMaps consider a simple
two dimensional integral over a unit hypercube with integrand:
def f(x):
return x[0] * x[1] ** 2
We want to create a grid that optimizes uniform Monte Carlo estimates
of the integral in y space. We do this by sampling the integrand
at a large number ny of random points y[j, d], where j=0...ny-1
and d=0,1, uniformly distributed throughout the integration
volume in y space. These samples be used to train the grid
using the following code:
import vegas
import numpy as np
def f(x):
return x[0] * x[1] ** 2
m = vegas.AdaptiveMap([[0, 1], [0, 1]], ninc=5)
ny = 1000
y = np.random.uniform(0., 1., (ny, 2)) # 1000 random y's
x = np.empty(y.shape, float) # work space
jac = np.empty(y.shape[0], float)
f2 = np.empty(y.shape[0], float)
print('intial grid:')
print(m.settings())
for itn in range(5): # 5 iterations to adapt
m.map(y, x, jac) # compute x's and jac
for j in range(ny): # compute training data
f2[j] = (jac[j] * f(x[j])) ** 2
m.add_training_data(y, f2) # adapt
m.adapt(alpha=1.5)
print('iteration %d:' % itn)
print(m.settings())
In each of the 5 iterations, the vegas.AdaptiveMap adjusts the
map, making increments smaller where f2 is larger and
larger where f2 is smaller.
The map converges after only 2 or 3 iterations, as
is clear from the output:
initial grid:
grid[ 0] = [ 0. 0.2 0.4 0.6 0.8 1. ]
grid[ 1] = [ 0. 0.2 0.4 0.6 0.8 1. ]
iteration 0:
grid[ 0] = [ 0. 0.412 0.62 0.76 0.883 1. ]
grid[ 1] = [ 0. 0.506 0.691 0.821 0.91 1. ]
iteration 1:
grid[ 0] = [ 0. 0.428 0.63 0.772 0.893 1. ]
grid[ 1] = [ 0. 0.531 0.713 0.832 0.921 1. ]
iteration 2:
grid[ 0] = [ 0. 0.433 0.63 0.772 0.894 1. ]
grid[ 1] = [ 0. 0.533 0.714 0.831 0.922 1. ]
iteration 3:
grid[ 0] = [ 0. 0.435 0.631 0.772 0.894 1. ]
grid[ 1] = [ 0. 0.533 0.715 0.831 0.923 1. ]
iteration 4:
grid[ 0] = [ 0. 0.436 0.631 0.772 0.895 1. ]
grid[ 1] = [ 0. 0.533 0.715 0.831 0.924 1. ]
The grid increments along direction 0 shrink at larger
values x[0], varying as 1/x[0]. Along direction 1
the increments shrink more quickly varying like 1/x[1]**2.
vegas samples the integrand in order to estimate the integral.
It uses those same samples to train its vegas.AdaptiveMap in this
fashion, for use in subsequent iterations of the algorithm.
- class vegas.AdaptiveMap¶
Adaptive map
y->x(y)for multidimensionalyandx.An
AdaptiveMapdefines a multidimensional mapy -> x(y)from the unit hypercube, with0 <= y[d] <= 1, to an arbitrary hypercube inxspace. Each direction is mapped independently with a Jacobian that is tunable (i.e., “adaptive”).The map is specified by a grid in
x-space that, by definition, maps into a uniformly spaced grid iny-space. The nodes of the grid are specified bygrid[d, i]where d is the direction (d=0,1...dim-1) andilabels the grid point (i=0,1...N). The mapping for a specific pointyintoxspace is:y[d] -> x[d] = grid[d, i(y[d])] + inc[d, i(y[d])] * delta(y[d])
where
i(y)=floor(y*N),delta(y)=y*N - i(y), andinc[d, i] = grid[d, i+1] - grid[d, i]. The Jacobian for this map,dx[d]/dy[d] = inc[d, i(y[d])] * N,
is piece-wise constant and proportional to the
x-space grid spacing. Each increment in thex-space grid maps into an increment of size1/Nin the correspondingyspace. So regions inxspace whereinc[d, i]is small are stretched out inyspace, while larger increments are compressed.The
xgrid for anAdaptiveMapcan be specified explicitly when the map is created: for example,m = AdaptiveMap([[0, 0.1, 1], [-1, 0, 1]])
creates a two-dimensional map where the
x[0]interval(0,0.1)and(0.1,1)map into they[0]intervals(0,0.5)and(0.5,1)respectively, whilex[1]intervals(-1,0)and(0,1)map intoy[1]intervals(0,0.5)and(0.5,1).More typically, an uniform map with
nincincrements is first created: for example,m = AdaptiveMap([[0, 1], [-1, 1]], ninc=1000)
creates a two-dimensional grid, with 1000 increments in each direction, that spans the volume
0<=x[0]<=1,-1<=x[1]<=1. This map is then trained with dataf[j]corresponding tonypointsy[j, d], withj=0...ny-1, (usually) uniformly distributed in y space: for example,m.add_training_data(y, f) m.adapt(alpha=1.5)
m.adapt(alpha=1.5)shrinks grid increments wheref[j]is large, and expands them wheref[j]is small. Usually one has to iterate over several sets ofys andfs before the grid has fully adapted.The speed with which the grid adapts is determined by parameter
alpha. Large (positive) values imply rapid adaptation, while small values (much less than one) imply slow adaptation. As in any iterative process that involves random numbers, it is usually a good idea to slow adaptation down in order to avoid instabilities caused by random fluctuations.- Parameters:
grid (list of arrays) – Initial
xgrid, wheregrid[d][i]is thei-th node in directiond. Different directions can have different numbers of nodes.ninc (int or array or
None) –ninc[d](orninc, if it is a number) is the number of increments along directiondin the newxgrid. The new grid is designed to give the same Jacobiandx(y)/dyas the original grid. The default value,ninc=None, leaves the grid unchanged.
- Attributes and methods:
- dim¶
Number of dimensions.
- ninc¶
ninc[d]is the number increments in directiond.
- grid¶
The nodes of the grid defining the maps are
self.grid[d, i]whered=0...specifies the direction andi=0...self.nincthe node.
- inc¶
The increment widths of the grid:
self.inc[d, i] = self.grid[d, i + 1] - self.grid[d, i]
- adapt(alpha=0.0, ninc=None)¶
Adapt grid to accumulated training data.
self.adapt(...)projects the training data onto each axis independently and maps it intoxspace. It shrinksx-grid increments in regions where the projected training data is large, and grows increments where the projected data is small. The grid along any direction is unchanged if the training data is constant along that direction.The number of increments along a direction can be changed by setting parameter
ninc(array or number).The grid does not change if no training data has been accumulated, unless
nincis specified, in which case the number of increments is adjusted while preserving the relative density of increments at different values ofx.- Parameters:
alpha (double) – Determines the speed with which the grid adapts to training data. Large (postive) values imply rapid evolution; small values (much less than one) imply slow evolution. Typical values are of order one. Choosing
alpha<0causes adaptation to the unmodified training data (usually not a good idea).ninc (int or array or None) – The number of increments in the new grid is
ninc[d](orninc, if it is a number) in directiond. The number is unchanged from the old grid ifnincis omitted (or equalsNone, which is the default).
- add_training_data(y, f, ny=-1)¶
Add training data
ffory-space pointsy.Accumulates training data for later use by
self.adapt(). Grid increments will be made smaller in regions wherefis larger than average, and larger wherefis smaller than average. The grid is unchanged (converged?) whenfis constant across the grid.- Parameters:
y (array) –
yvalues corresponding to the training data.yis a contiguous 2-d array, wherey[j, d]is for points along directiond.f (array) – Training function values.
f[j]corresponds to pointy[j, d]iny-space.ny (int) – Number of
ypoints:y[j, d]ford=0...dim-1andj=0...ny-1.nyis set toy.shape[0]if it is omitted (or negative).
- adapt_to_samples(x, f, nitn=5, alpha=1.0, nproc=1)¶
Adapt map to data
{x, f(x)}.Replace grid with one that is optimized for integrating function
f(x). New grid is found iteratively- Parameters:
x (array) –
x[:, d]are the components of the sample points in directiond=0,1...self.dim-1.f (callable or array) – Function
f(x)to be adapted to. Iffis an array, it is assumes to contain valuesf[i]corresponding to the function evaluated at pointsx[i].nitn (int) – Number of iterations to use in adaptation. Default is
nitn=5.alpha (float) – Damping parameter for adaptation. Default is
alpha=1.0. Smaller values slow the iterative adaptation, to improve stability of convergence.nproc (int or None) –
Number of processes/processors to use. If
nproc>1Python’smultiprocessingmodule is used to spread the calculation across multiple processors. There is a significant overhead involved in using multiple processors so this option is useful mainly when very high dimenions or large numbers of samples are involved. When using themultiprocessingmodule in its default mode for MacOS and Windows, it is important that the main module can be safely imported (i.e., without launching new processes). This can be accomplished with some version of theif __name__ == '__main__`:construct in the main module: e.g.,if __name__ == '__main__': main()
This is not an issue on other Unix platforms. See the
multiprocessingdocumentation for more information. Setnproc=Noneto use all the processors on the machine (equivalent tonproc=os.cpu_count()). Default value isnproc=1. (Requires Python 3.3 or later.)
- __call__(y)¶
Return
xvalues corresponding toy.ycan be a singledim-dimensional point, or it can be an arrayy[i,j, ..., d]of such points (d=0..dim-1).If
y=None(default),yis set equal to a (uniform) random point in the volume.
- jac(y)¶
Return the map’s Jacobian at
y.ycan be a singledim-dimensional point, or it can be an arrayy[i,j,...,d]of such points (d=0..dim-1). Returns an arrayjacwherejac[i,j,...]is the (multidimensional) Jacobian (dx/dy) corresponding toy[i,j,...].
- jac1d(y)¶
Return the map’s Jacobian at
yfor each direction.ycan be a singledim-dimensional point, or it can be an arrayy[i,j,...,d]of such points (d=0..dim-1). Returns an arrayjacwherejac[i,j,...,d]is the (one-dimensional) Jacobian (dx[d]/dy[d]) corresponding toy[i,j,...,d].
- make_uniform(ninc=None)¶
Replace the grid with a uniform grid.
The new grid has
ninc[d](orninc, if it is a number) increments along each direction ifnincis specified. Ifninc=None(default), the new grid has the same number of increments in each direction as the old grid.
- map(y, x, jac, ny=-1)¶
Map y to x, where jac is the Jacobian (
dx/dy).y[j, d]is an array ofnyy-values for directiond.x[j, d]is filled with the correspondingxvalues, andjac[j]is filled with the corresponding Jacobian values.xandjacmust be preallocated: for example,x = numpy.empty(y.shape, float) jac = numpy.empty(y.shape[0], float)
- Parameters:
y (array) –
yvalues to be mapped.yis a contiguous 2-d array, wherey[j, d]contains values for points along directiond.x (array) – Container for
x[j, d]values corresponding toy[j, d]. Must be a contiguous 2-d array.jac (array) – Container for Jacobian values
jac[j](= dx/dy) corresponding toy[j, d]. Must be a contiguous 1-d array.ny (int) – Number of
ypoints:y[j, d]ford=0...dim-1andj=0...ny-1.nyis set toy.shape[0]if it is omitted (or negative).
- invmap(x, y, jac, nx=-1)¶
Map x to y, where jac is the Jacobian (
dx/dy).y[j, d]is an array ofnyy-values for directiond.x[j, d]is filled with the correspondingxvalues, andjac[j]is filled with the corresponding Jacobian values.xandjacmust be preallocated: for example,x = numpy.empty(y.shape, float) jac = numpy.empty(y.shape[0], float)
- Parameters:
x (array) –
xvalues to be mapped toy-space.xis a contiguous 2-d array, wherex[j, d]contains values for points along directiond.y (array) – Container for
y[j, d]values corresponding tox[j, d]. Must be a contiguous 2-d arrayjac (array) – Container for Jacobian values
jac[j](= dx/dy) corresponding toy[j, d]. Must be a contiguous 1-d arraynx (int) – Number of
xpoints:x[j, d]ford=0...dim-1andj=0...nx-1.nxis set tox.shape[0]if it is omitted (or negative).
- show_grid(ngrid=40, shrink=False)¶
Display plots showing the current grid.
- Parameters:
ngrid (int) – The number of grid nodes in each direction to include in the plot. The default is 40.
axes – List of pairs of directions to use in different views of the grid. Using
Nonein place of a direction plots the grid for only one direction. Omittingaxescauses a default set of pairings to be used.shrink – Display entire range of each axis if
False; otherwise shrink range to include just the nodes being displayed. The default isFalse.plotter –
matplotlibplotter to use for plots; plots are not displayed if set. Ignored ifNone, and plots are displayed usingmatplotlib.pyplot.
- settings(ngrid=5)¶
Create string with information about grid nodes.
Creates a string containing the locations of the nodes in the map grid for each direction. Parameter
ngridspecifies the maximum number of nodes to print (spread evenly over the grid).
- extract_grid()¶
Return a list of lists specifying the map’s grid.
- clear()¶
Clear information accumulated by
AdaptiveMap.add_training_data().
PDFIntegrator Objects¶
Expectation values using probability density functions defined by
collections of Gaussian random variables (see gvar)
can be evaluated using the following
specialized integrator:
- class vegas.PDFIntegrator(g, limit=1e15, scale=1., svdcut=1e-15)¶
vegasintegrator for PDF expectation values.PDFIntegrator(param, pdf)creates avegasintegrator that evaluates expectation values of arbitrary functionsf(p)with respect to the probability density functionpdf(p), wherepis a point in the parameter space defined byparam.paramis a collection ofgvar.GVars (Gaussian random variables) that together define a multi-dimensional Gaussian distribution with the same parameter space as the distribution described bypdf(p).PDFIntegratorinternally re-expresses the integrals over these parameters in terms of new variables that emphasize the region defined byparam(i.e., the region where the PDF associated with theparam’s Gaussian distribution is large). The new variables are also aligned with the principal axes ofparam’s correlation matrix, to facilitate integration.param’s means and covariances are chosen to emphasize the important regions of thepdf’s distribution (e.g.,parammight be set equal to the prior in a Bayesian analysis).paramis used to define and optimize the integration variables; it does not affect the values of the integrals but can have a big effect on the accuracy.The Gaussian PDF associated with
paramis used ifpdfis unspecified (i.e.,pdf=None, which is the default).Typical usage is illustrated by the following code, where dictionary
gspecifies both the parameterization (param) and the PDF:import vegas import gvar as gv import numpy as np g = gv.BufferDict() g['a'] = gv.gvar([10., 2.], [[1, 1.4], [1.4, 2]]) g['fb(b)'] = gv.BufferDict.uniform('fb', 2.9, 3.1) g_ev = vegas.PDFIntegrator(g) def f(p): a = p['a'] b = p['b'] return a[0] + np.fabs(a[1]) ** b result = g_ev(f, neval=10_000, nitn=5) print('<f(p)> =', result)
Here
gindicates a three-dimensional distribution where the first two variablesg['a'][0]andg['a'][1]are Gaussian with means 10 and 2, respectively, and covariance matrix [[1, 1.4], [1.4, 2.]]. The last variableg['b']is uniformly distributed on interval [2.9, 3.1]. The result is:<f(p)> = 30.145(83).PDFIntegratorevaluates integrals of bothf(p) * pdf(p)andpdf(p). The expectation value off(p)is the ratio of these two integrals (sopdf(p)need not be normalized). The result of aPDFIntegratorintegration has an extra attribute,result.pdfnorm, which is thevegasestimate of the integral over the PDF.- Parameters:
param – A
gvar.GVar, array ofgvar.GVars, or dictionary, whose values aregvar.GVars or arrays ofgvar.GVars, that specifies the integration parameters. When parameterpdf=None, the PDF is set equal to the Gaussian distribution corresponding toparam.pdf – The probability density function
pdf(p). The PDF’s parametersphave the same layout asparam(arrays or dictionary), with the same keys and/or shapes. The Gaussian PDF associated withparamis used whenpdf=None(default). Note that PDFs need not be normalized.adapt_to_pdf (bool) –
vegasadapts to the PDF whenadapt_to_pdf=True(default).vegasadapts topdf(p) * f(p)when calculating the expectation value off(p)ifadapt_to_pdf=False.limit (positive float) – Integration variables are determined from
param.limitlimits the range of each variable to a region of sizelimittimes the standard deviation on either side of the mean, where means and standard deviations are specified byparam. This can be useful if the functions being integrated misbehave for large parameter values (e.g.,numpy.expoverflows for a large range of arguments). Default islimit=100; results should become independent oflimitas it is increased.scale (positive float) – The integration variables are rescaled to emphasize parameter values of order
scaletimes the standard deviation measured from the mean, where means and standard deviations are specified byparam. The rescaling does not change the value of the integral but it can reduce uncertainties in thevegasestimate. Default isscale=1.0.svdcut (non-negative float or None) – If not
None, replace correlation matrix ofparamwith a new matrix whose small eigenvalues are modified: eigenvalues smaller thansvdcuttimes the maximum eigenvalueeig_maxare replaced bysvdcut*eig_max. This can ameliorate problems caused by roundoff errors when inverting the covariance matrix. It increases the uncertainty associated with the modified eigenvalues and so is conservative. Settingsvdcut=Noneorsvdcut=0leaves the covariance matrix unchanged. Default issvdcut=1e-12.
All other keyword parameters are passed on to the the underlying
vegas.Integrator; theuses_jackeyword is ignored.- Methods:
- __call__(f=None, pdf=None, adapt_to_pdf=None, save=None, saveall=None, **kargs)¶
Estimate expectation value of function
f(p).Uses module
vegasto estimate the integral off(p)multiplied by the probability density function associated withg(i.e.,pdf(p)). At the same time it integrates the PDF. The ratio of the two integrals is the expectation value.- Parameters:
f (function) – Function
f(p)to integrate. Integral is the expectation value of the function with respect to the distribution. The function can return a number, an array of numbers, or a dictionary whose values are numbers or arrays of numbers. Settingf=Nonemeans that only the PDF is integrated. Integrals can be substantially faster iff(p)(andpdf(p)if set) are batch functions (seevegasdocumentation).pdf – If specified,
pdf(p)is used as the probability density function rather than the Gaussian PDF associated withg. The Gaussian PDF is used ifpdf=None(default). Note that PDFs need not be normalized.adapt_to_pdf (bool) –
vegasadapts to the PDF whenadapt_to_pdf=True(default).vegasadapts topdf(p) * f(p)ifadapt_to_pdf=False.save (str or file or None) –
Writes
resultsinto pickle file specified bysaveat the end of each iteration. For example, settingsave='results.pkl'means that the results returned by the last vegas iteration can be reconstructed later using:import pickle with open('results.pkl', 'rb') as ifile: results = pickle.load(ifile)
Ignored if
save=None(default).saveall (str or file or None) –
Writes
(results, integrator)into pickle file specified bysaveallat the end of each iteration. For example, settingsaveall='allresults.pkl'means that the results returned by the last vegas iteration, together with a clone of the (adapted) integrator, can be reconstructed later using:import pickle with open('allresults.pkl', 'rb') as ifile: results, integrator = pickle.load(ifile)
Ignored if
saveall=None(default).
All other keyword arguments are passed on to a
vegasintegrator; see thevegasdocumentation for further information.- Returns:
Expectation value(s) of
f(p)as object of typevegas.PDFEV,vegas.PDFEVArray, orvegas.PDFEVDict.
- stats(f, moments=False, histograms=False, **kargs)¶
Statistical analysis of function
f(p).Uses the
vegasintegrator to evaluate the expectation values and (co)variances off(p)with respect to the probability density function associated with thePDFIntegrator. Typical usage is illustrated by:>>> import gvar as gv >>> import vegas >>> g = gv.gvar(dict(a='1.0(5)', b='2(1)')) * gv.gvar('1.0(5)') >>> g_ev = vegas.PDFIntegrator(g) >>> g_ev(neval=10_000) # adapt the integrator to the PDF >>> @vegas.rbatchintegrand ... def f(p): ... fp = dict(a=p['a'], b=p['b']) ... fp['a**2 * b'] = p['a']**2 * p['b'] ... return fp >>> r = g_ev.stats(f) >>> print(r) {'a': 1.00(71), 'b': 2.0(1.4), 'a**2 * b': 4.0(6.1)} >>> print(r.vegas_mean['a**2 * b']) 3.9972(30) >>> print(r.vegas_cov['a**2 * b', 'a**2 * b'] ** 0.5) 6.073(13)
g_ev.stats(f)returns a dictionary ofgvar.GVars whose means and (co)variances are calculated from integrals off(p) * pdf(p)andf(p)**2 * pdf(p), wherepdf(p)is the probability density function associated withg. The means and standard deviations for each component off(p)are displayed byprint(r). The values for the means and standard deviations have uncertainties coming from the integrations (vegaserrors) but these are negligible compared to the standard deviations. (The last two print statements show thevegasresults for the mean and standard deviation inr['a**2 * b']: 3.9972(30) and 6.073(13), respectively.)Th Gaussian approximation for the expectation value of
f(p)is given by>>> print(f(g)) {'a': 1.00(71), 'b': 2.0(1.4), 'a**2 * b': 2.0(3.7)}
Results for
aandbagree with the results fromg_ev.stats(f), as expected since the distributions for these quantities are (obviously) Gaussian. Results fora**2 * b, however, are quite different, indicating a distribution that is not Gaussian.Additional statistical data are collected by setting keywords
moments=Trueand/orhistogram=True:>>> r = g_ev.stats(f, moments=True, histograms=True) >>> for k in r: ... print(10 * '-', k) ... print(r.stats[k]) ---------- a mean = 0.99972(23) sdev = 0.70707(29) skew = -0.0036(20) ex_kurt = -0.0079(49) split-normal: 1.0013(14) +/- 0.70862(97)/0.71091(98) median: 0.99927(62) +/- 0.7077(10)/0.7063(10) ---------- b mean = 1.99954(47) sdev = 1.41424(72) skew = -0.0041(28) ex_kurt = -0.0074(65) split-normal: 2.0042(33) +/- 1.4162(23)/1.4224(24) median: 1.9977(11) +/- 1.4162(18)/1.4115(19) ---------- a**2 * b mean = 3.9957(29) sdev = 6.054(12) skew = 3.048(22) ex_kurt = 14.52(35) split-normal: -0.4891(25) +/- 6.9578(88)/0.519(10) median: 1.7447(24) +/- 6.284(12)/2.0693(26)
where the uncertainties are all
vegaserrors. Here the integrator was used to calculate the first four moments of the distributions for each component off(p), from which the mean, standard deviation, skewness, and excess kurtosis of those distributions are calculated. As expected the first two distribuitons here are clearly Gaussian, but the distribution fora**2 * bis not.The integrator also calculates histograms for each of the distributions and fits them to two different two-sided Gaussians: one is a continuous split-normal distribution, and the other is centered on the median of the distribution and is discontinuous there. (For more information see the documentation for
gvar.PDFStatistics.) Both models suggest large asymmetries in the distribution fora**2 * b. The histogram for this distribution can be displayed using:>>> r.stats['a**2 * b'].plot_histogram(show=True)
Note that
vegasadaptation is turned off (adapt=False) by default inPDFIntegrator.stats(). This setting can be overridden by setting theadaptparameter explicitly, but this is not recommended.- Parameters:
f (callable) – Statistics are calculated for the components of the output from function
f(p), wherepis a point drawn from the distribution specified by theparamorpdfassociated with thePDFIntegrator. Parametersphave the same structure asparam(i.e., array or dictionary). Iff=None, it is replaced byf=lbatchintegrand(lambda p:p).moments (bool) – If
True, moments are calculated so that the skewness and excess kurtosis can be determined.histograms (bool or dict) –
Setting
histograms=Truecauses histograms to be calculated for the distributions associated with each component of the output fromf(p). Alternatively,histogramscan be set equal to a dictionary to specify the the widthbinwidthof each histogram bin, the total numbernbinof bins, and/or the locationlocof each histogram: for example,histograms=dict( binwidth=0.3, nbin=30, loc=gv.gvar({ 'a': '1.0(5)', 'b': '2(1)', 'a**2 * b': '2.5(2.7)' }), )
where
locspecifies the location of the center of the histogram for each output quantity (e.g.,loc['a'].mean) and the width of the bins (e.g.,binwidth * loc['a'].sdev). Iflocis not specified explicitly, it is determined from a simulation using values drawn from the Gaussian distribution forself.g(or from the distribution described byself.pdfif it is specified).kargs (dict) – Additional keywords passed on to the integrator.
- Returns:
Expectation value(s) of
f(p)as an object of typevegas.PDFEV,vegas.PDFEVArray, orvegas.PDFEVDict.
- sample(nbatch, mode='rbatch')¶
Generate random samples from the integrator’s PDF.
Typical usage is:
import gvar as gv import numpy as np import vegas @vegas.rbatchintegrand def g_pdf(p): ans = 0 h = 1. for p0 in [0.3, 0.6]: ans += h * np.exp(-np.sum((p-p0)**2, axis=0)/2/.01) h /= 2 return ans g_param = gv.gvar([0.5, 0.5], [[.25, .2], [.2, .25]]) g_ev = vegas.PDFIntegrator(param=g_param, pdf=g_pdf) # adapt integrator to g_pdf(p) and evaluate <p> g_ev(neval=4000, nitn=10) r = g_ev.stats() print('<p> =', r, '(vegas)') # sample g_pdf(p) wgts, p_samples = g_ev.sample(nbatch=40_000) # evaluate mean values <p> and <cos(p0)> p_avg = np.sum(wgts * p_samples, axis=1) cosp0_avg = np.sum(wgts * np.cos(p_samples[0])) print('<p> =', p_avg, '(sample)') print('<cos(p0)> =', cosp0_avg, '(sample)')
Here
p_samples[d, i]is a batch of about 40,000 random samples for parameterp[d]drawn from the (bimodal) distribution with PDFg_pdf(p). Indexd=0,1labels directions in parameter space, while indexilabels the sample. The samples are weighted bywgts[i]; the sum of all weights equals one. The batch index inp_samplesis the rightmost index because by defaultmode='rbatch'. (Setmode='lbatch'to move the batch index to the leftmost position:p_samples[i, d].) The output from this script is:<p> = [0.40(17) 0.40(17)] (vegas) <p> = [0.40011804 0.39999454] (sample) <cos(p0)> = 0.9074221724843065 (sample)
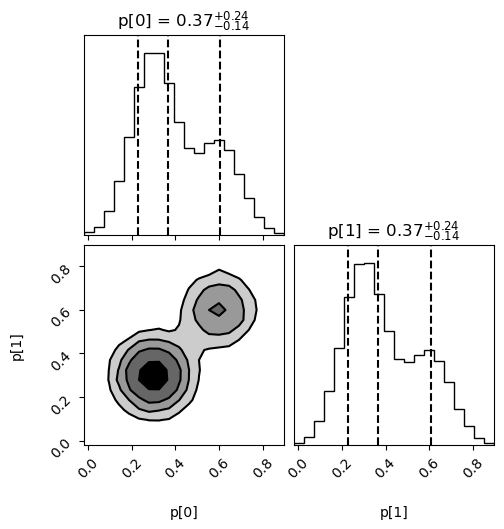
Samples are also useful for making histograms and contour plots of the probability density. For example, the following code uses the
cornerPython module to create histograms for each parameter, and a contour plot showing their joint distribution:import corner import matplotlib.pyplot as plt corner.corner( data=p_samples.T, weights=wgts, labels=['p[0]', 'p[1]'], range=[0.999, 0.999], show_titles=True, quantiles=[0.16, 0.5, 0.84], ) plt.show()
The output, showing the bimodal structure, is:
- Parameters:
nbatch (int) – The integrator will return at least
nbatchsamples drawn from its PDF. The actual number of samples is the smallest multiple ofself.last_nevalthat is equal to or larger thannbatch. Results are packaged in arrays or dictionaries whose elements have an extra index labeling the different samples in the batch. The batch index is the rightmost index ifmode='rbatch'; it is the leftmost index ifmodeis'lbatch'.mode (bool) – Batch mode. Allowed modes are
'rbatch'or'lbatch', corresponding to batch indices that are on the right or the left, respectively. Default ismode='rbatch'.
- Returns:
A tuple
(wgts,samples)containing samples drawn from the integrator’s PDF, together with their weightswgts. The weighted sample points are distributed through parameter space with a density proportional to the PDF.In general,
samplesis either a dictionary or an array depending upon the format ofvegas.PDFIntegratorparameterparam. For example, ifparam = gv.gvar(dict(s='1.5(1)', v=['3.2(8)', '1.1(4)']))
then
samples['s'][i]is a sample for parameterp['s']where indexi=0,1...nbatch(approx)labels the sample. The corresponding sample forp['v'][d], whered=0or1, issamples['v'][d, i]providedmode='rbatch', which is the default. (Otherwise it isp['v'][i, d], formode='lbatch'.) The corresponding weight for this sample iswgts[i].When
paramis an array,samplesis an array with the same shape plus an extra sample index which is either on the right (mode='rbatch', default) or left (mode='lbatch').
Other Objects and Functions¶
- class vegas.RAvg(weighted=True, itn_results=None, sum_neval=0, _rescale=True)¶
Running average of scalar-valued Monte Carlo estimates.
This class accumulates independent Monte Carlo estimates (e.g., of an integral) and combines them into a single average. It is derived from
gvar.GVar(from thegvarmodule if it is present) and represents a Gaussian random variable.Different estimates are weighted by their inverse variances if parameter
weight=True; otherwise straight, unweighted averages are used.- Attributes and methods:
- mean¶
The mean value of the weighted average.
- sdev¶
The standard deviation of the weighted average.
- chi2¶
chi**2 of weighted average.
- dof¶
Number of degrees of freedom in weighted average.
- Q¶
Q or p-value of weighted average’s chi**2.
- itn_results¶
A list of the results from each iteration.
- sum_neval¶
Total number of integrand evaluations used in all iterations.
- add(g)¶
Add estimate
gto the running average.
- class vegas.RAvgArray(shape=None, dtype=<class 'object'>, buffer=None, offset=0, strides=None, order=None, weighted=True, itn_results=None, sum_neval=0, rescale=True)¶
Running average of array-valued Monte Carlo estimates.
This class accumulates independent arrays of Monte Carlo estimates (e.g., of an integral) and combines them into an array of averages. It is derived from
numpy.ndarray. The array elements aregvar.GVars (from thegvarmodule if present) and represent Gaussian random variables.Different estimates are weighted by their inverse covariance matrices if parameter
weight=True; otherwise straight, unweighted averages are used.- Attributes and methods:
- chi2¶
chi**2 of weighted average.
- dof¶
Number of degrees of freedom in weighted average.
- Q¶
Q or p-value of weighted average’s chi**2.
- itn_results¶
A list of the results from each iteration.
- sum_neval¶
Total number of integrand evaluations used in all iterations.
- add(g)¶
Add estimate
gto the running average.
- summary(extended=False, weighted=None)¶
Assemble summary of results, iteration-by-iteration, into a string.
- Parameters:
extended (bool) – Include a table of final averages for every component of the integrand if
True. Default isFalse.weighted (bool) – Display weighted averages of results from different iterations if
True; otherwise show unweighted averages. Default behavior is determined byvegas.
- class vegas.RAvgDict(dictionary=None, weighted=True, itn_results=None, sum_neval=0, rescale=True)¶
Running average of dictionary-valued Monte Carlo estimates.
This class accumulates independent dictionaries of Monte Carlo estimates (e.g., of an integral) and combines them into a dictionary of averages. It is derived from
gvar.BufferDict. The dictionary values aregvar.GVars or arrays ofgvar.GVars.Different estimates are weighted by their inverse covariance matrices if parameter
weight=True; otherwise straight, unweighted averages are used.- Attributes and methods:
- chi2¶
chi**2 of weighted average.
- dof¶
Number of degrees of freedom in weighted average.
- Q¶
Q or p-value of weighted average’s chi**2.
- itn_results¶
A list of the results from each iteration.
- sum_neval¶
Total number of integrand evaluations used in all iterations.
- add(g)¶
Augment buffer with data
v, indexed by keyk.vis either a scalar or anumpyarray (or a list or other data type that can be changed into a numpy.array). Ifvis anumpyarray, it can have any shape.Same as
self[k] = vexcept whenkis already used inself, in which case aValueErroris raised.
- summary(extended=False, weighted=None)¶
Assemble summary of results, iteration-by-iteration, into a string.
- Parameters:
extended (bool) – Include a table of final averages for every component of the integrand if
True. Default isFalse.weighted (bool) – Display weighted averages of results from different iterations if
True; otherwise show unweighted averages. Default behavior is determined byvegas.
- class vegas.PDFEV(results, analyzer=None)¶
Expectation value from
vegas.PDFIntegrator.Expectation values are returned by
vegas.PDFIntegrator.__call__()andvegas.PDFIntegrator.stats:>>> g = gvar.gvar(['1(1)', '10(10)']) >>> g_ev = vegas.PDFIntegrator(g) >>> def f(p): ... return p[0] * p[1] >>> print(g_ev(f)) 10.051(57) >>> print(g_ev.stats(f)) 10(17)
In the first case, the quoted error is the uncertainty in the
vegasestimate of the mean off(p). In the second case, the quoted uncertainty is the standard deviation evaluated with respect to the Gaussian distribution associated withg(added in quadrature to thevegaserror, which is negligible here).vegas.PDFEVs have the following attributes:- pdfnorm¶
Divide PDF by
pdfnormto normalize it.
- results¶
Results from the underlying integrals.
- Type:
In addition, they have all the attributes of the
vegas.RAvgDict(results) corresponding to the underlying integrals.A
vegas.PDFEVreturned byvegas.PDFIntegrator.stats(self, f...)has three further attributes:- stats¶
An instance of
gvar.PDFStatisticscontaining statistical information about the distribution off(p).
- class vegas.PDFEVArray(results, analyzer=None)¶
Array of expectation values from
vegas.PDFIntegrator.Expectation values are returned by
vegas.PDFIntegrator.__call__()andvegas.PDFIntegrator.stats:>>> g = gvar.gvar(['1(1)', '10(10)']) >>> g_ev = vegas.PDFIntegrator(g) >>> def f(p): ... return [p[0], p[1], p[0] * p[1]] >>> print(g_ev(f)) [0.9992(31) 10.024(29) 10.051(57)] >>> print(g_ev.stats(f)) [1.0(1.0) 10(10) 10(17)]
In the first case, the quoted errors are the uncertainties in the
vegasestimates of the means. In the second case, the quoted uncertainties are the standard deviations evaluated with respect to the Gaussian distribution associated withg(added in quadrature to thevegaserrors, which are negligible here).vegas.PDFEVArrays have the following attributes:- pdfnorm¶
Divide PDF by
pdfnormto normalize it.
- results¶
Results from the underlying integrals.
- Type:
In addition, they have all the attributes of the
vegas.RAvgDict(results) corresponding to the underlying integrals.A
vegas.PDFEVArraysreturned byvegas.PDFIntegrator.stats(self, f...)has three further attributes:- stats¶
s.stats[i]is agvar.PDFStatisticsobject containing statistical information about the distribution off(p)[i].
- class vegas.PDFEVDict(results, analyzer=None)¶
Dictionary of expectation values from
vegas.PDFIntegrator.Expectation values are returned by
vegas.PDFIntegrator.__call__()andvegas.PDFIntegrator.stats:>>> g = gvar.gvar(['1(1)', '10(10)']) >>> g_ev = vegas.PDFIntegrator(g) >>> def f(p): ... return dict(p=p, prod=p[0] * p[1]) >>> print(g_ev(f)) {'p': array([0.9992(31), 10.024(29)], dtype=object), 'prod': 10.051(57)} >>> print(g_ev.stats(f)) {'p': array([1.0(1.0), 10(10)], dtype=object), 'prod': 10(17)}
In the first case, the quoted errors are the uncertainties in the
vegasestimates of the means. In the second case, the quoted uncertainties are the standard deviations evaluated with respect to the Gaussian distribution associated withg(added in quadrature to thevegaserrors, which are negligible here).vegas.PDFEVDictobjects have the following attributes:- pdfnorm¶
Divide PDF by
pdfnormto normalize it.
- results¶
Results from the underlying integrals.
- Type:
In addition, they have all the attributes of the
vegas.RAvgDict(results) corresponding to the underlying integrals.A
vegas.PDFEVDictobjectsreturned byvegas.PDFIntegrator.stats()has three further attributes:- stats¶
s.stats[k]is agvar.PDFStatisticsobject containing statistical information about the distribution off(p)[k].
- vegas.lbatchintegrand(f)¶
Decorator for batch integrand functions.
Applying
vegas.lbatchintegrand()to a functionfcnrepackages the function in a format thatvegascan understand. Appropriate functions take anumpyarray of integration pointsx[i, d]as an argument, wherei=0...labels the integration point andd=0...labels direction, and return an arrayf[i]of integrand values (or arrays of integrand values) for the corresponding points. The meaning offcn(x)is unchanged by the decorator.An example is
import vegas import numpy as np @vegas.lbatchintegrand # or @vegas.lbatchintegrand def f(x): return np.exp(-x[:, 0] - x[:, 1])
for the two-dimensional integrand
. When integrands have dictionary arguments
xd, each element of the dictionary has an extra index (on the left):xd[key][:, ...].vegas.batchintegrand()is the same asvegas.lbatchintegrand().
- class vegas.LBatchIntegrand¶
Wrapper for lbatch integrands.
Used by
vegas.lbatchintegrand().vegas.LBatchIntegrandis the same asvegas.BatchIntegrand.
- vegas.rbatchintegrand(f)¶
Same as
vegas.lbatchintegrand()but with batch indices on the right (not left).
- class vegas.RBatchIntegrand¶
Same as
vegas.LBatchIntegrandbut with batch indices on the right (not left).
- vegas.ravg(reslist, weighted=None, rescale=None)¶
Create running average from list of
vegasresults.This function is used to change how the weighted average of
vegasresults is calculated. For example, the following code discards the first five results (wherevegasis still adapting) and does an unweighted average of the last five:import vegas def fcn(p): return p[0] * p[1] * p[2] * p[3] * 16. itg = vegas.Integrator(4 * [[0,1]]) r = itg(fcn) print(r.summary()) ur = vegas.ravg(r.itn_results[5:], weighted=False) print(ur.summary())
The unweighted average can be useful because it is unbiased. The output is:
itn integral wgt average chi2/dof Q ------------------------------------------------------- 1 1.013(19) 1.013(19) 0.00 1.00 2 0.997(14) 1.002(11) 0.45 0.50 3 1.021(12) 1.0112(80) 0.91 0.40 4 0.9785(97) 0.9980(62) 2.84 0.04 5 1.0067(85) 1.0010(50) 2.30 0.06 6 0.9996(75) 1.0006(42) 1.85 0.10 7 1.0020(61) 1.0010(34) 1.54 0.16 8 1.0051(52) 1.0022(29) 1.39 0.21 9 1.0046(47) 1.0029(24) 1.23 0.27 10 0.9976(47) 1.0018(22) 1.21 0.28 itn integral average chi2/dof Q ------------------------------------------------------- 1 0.9996(75) 0.9996(75) 0.00 1.00 2 1.0020(61) 1.0008(48) 0.06 0.81 3 1.0051(52) 1.0022(37) 0.19 0.83 4 1.0046(47) 1.0028(30) 0.18 0.91 5 0.9976(47) 1.0018(26) 0.31 0.87
- Parameters:
reslist (list) – List whose elements are
gvar.GVars, arrays ofgvar.GVars, or dictionaries whose values aregvar.GVars or arrays ofgvar.GVars. Alternativelyreslistcan be the object returned by a call to avegas.Integratorobject (i.e, an instance of any ofvegas.RAvg,vegas.RAvgArray,vegas.RAvgArray,vegas.PDFEV,vegas.PDFEVArray,vegas.PDFEVArray).weighted (bool) – Running average is weighted (by the inverse covariance matrix) if
True. Otherwise the average is unweighted, which makes most sense ifreslistitems were generated byvegaswith little or no adaptation (e.g., withadapt=False). Ifweightedis not specified (or isNone), it is set equal togetattr(reslist, 'weighted', True).rescale – Integration results are divided by
rescalebefore taking the weighted average ifweighted=True; otherwiserescaleis ignored. Settingrescale=Trueis equivalent to settingrescale=reslist[-1]. Ifrescaleis not specified (or isNone), it is set equal togetattr(reslist, 'rescale', True).